Overview of MLCPP methodology
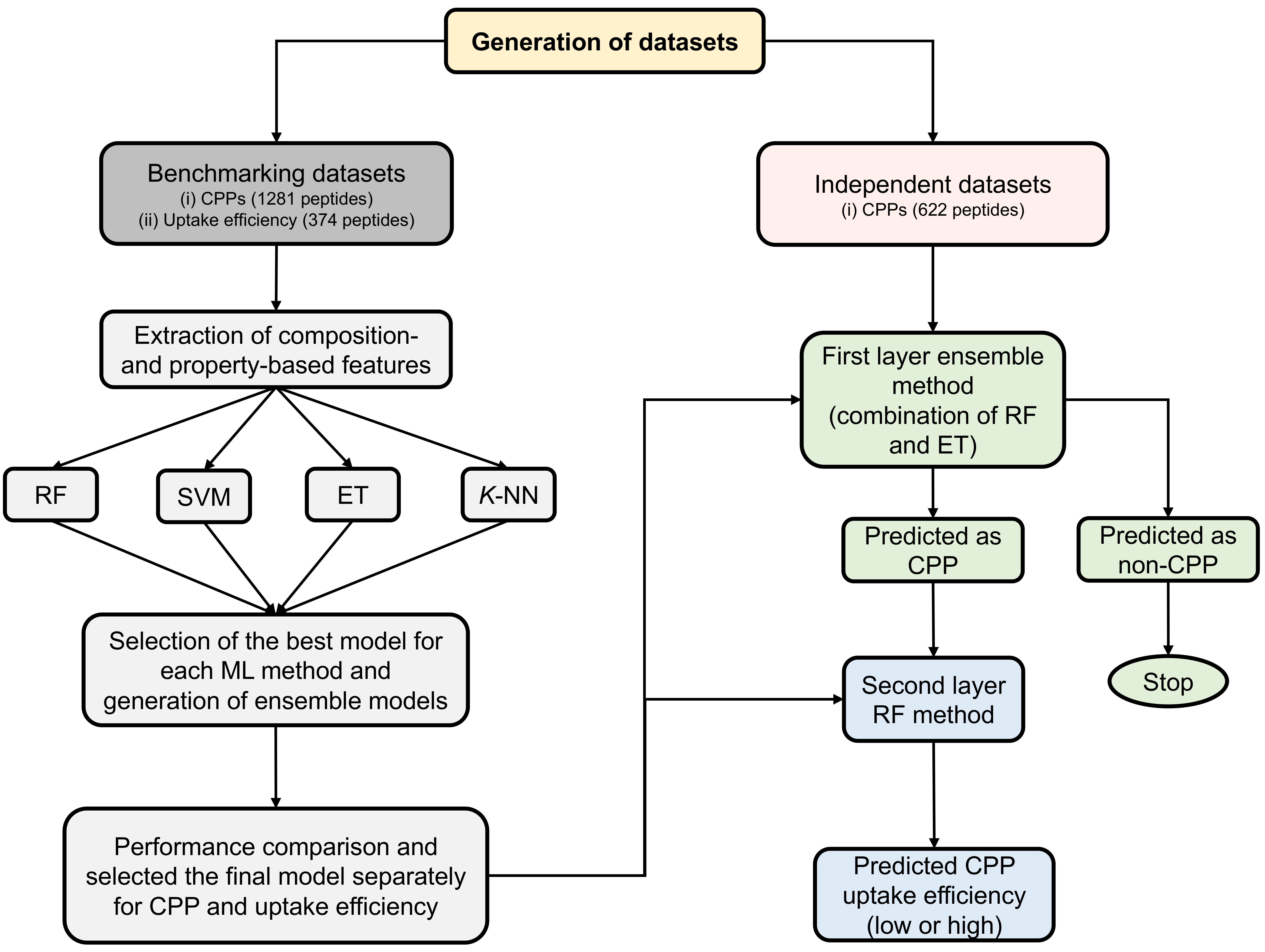
It shows the four stages of development of our tools. The first stage involved dataset generation. In the second stage, different features were extracted from the peptide sequences, including amino acid composition (AAC), atomic composition (ATC), composition-transition-distribution (CTD), dipeptide composition (DPC), and physicochemical properties (PCP). In the third stage, construction of the four-different machine-learning (ML)-based classifiers including random forest (RF), support vector machine (SVM), extra tree classifier (ET) and k-nearest neighbor (k-NN) using various feature sets and a selection of the best model for each ML method. We compared the performance of the individual method and ensemble methods, and constructed the first-layer prediction model (CPP or non-CPP) and the second-layer prediction model (low or high uptake efficiency).