Overview of iBCE-EL methodology
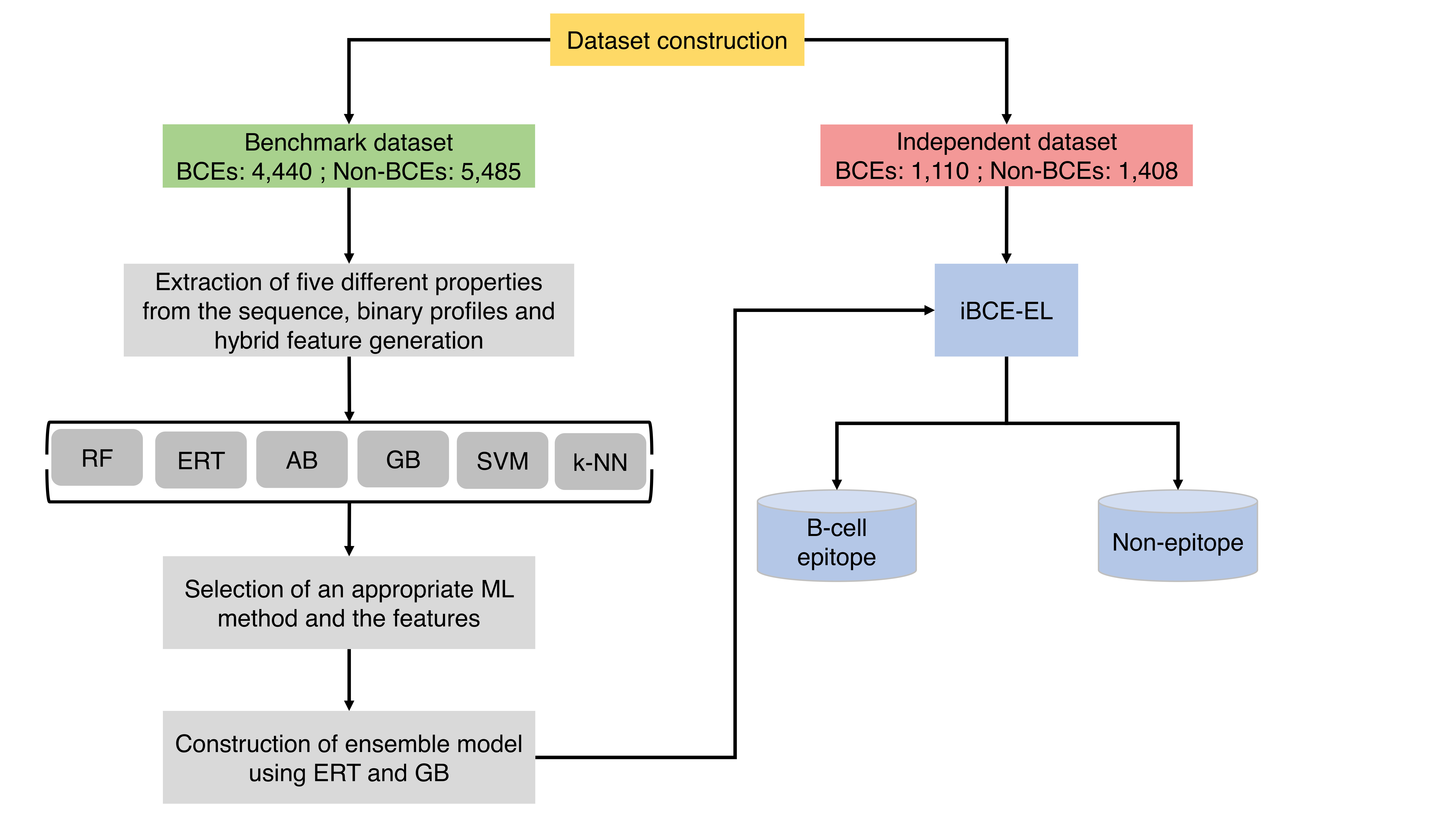
A flowchart describing iBCE-EL is illustrated above, which consists of the following 4 stages: (1) construction of a nr benchmarking dataset of 9,925 peptides (4,440 BCEs and 5,485 non-BCEs) and an independent dataset of 2,518 peptides (1,110 BCEs and 1,408 non-BCEs) from IEDB; (2) extraction of various features from peptide sequences including AAC, AAI, CTD, DPC and PCP, the generation of hybrid features (various combination of individual composition) and binary profiles; (3) exploration of six different ML algorithms and selection of an appropriate algorithms and the corresponding features; and (4) construction of ensemble model.